GNE: A deep learning framework for gene network inference by aggregating biological information
This is the TensorFlow implementation of the GNE as described in our paper. The code was forked initially from here.
WE provide PyTorch implementation for GNE that encodes network struture only in BionetEmbedding. Note that this is a minimal version of this TensorFlow implementation.
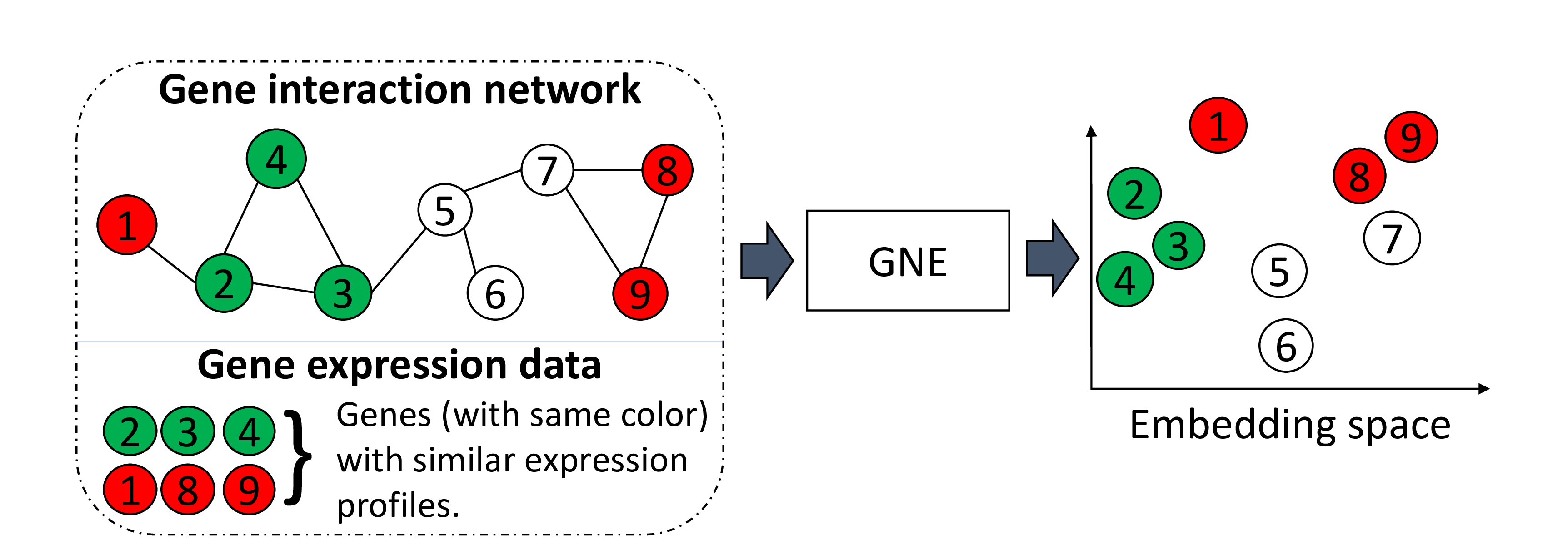
GNE integrates gene interaction network with gene expression data to learn a more informative representations for gene network, which can be plugged into off-the-shelf machine learning methods for diverse functional inference tasks: gene function prediction, gene ontology reconstruction, and genetic interaction prediction.
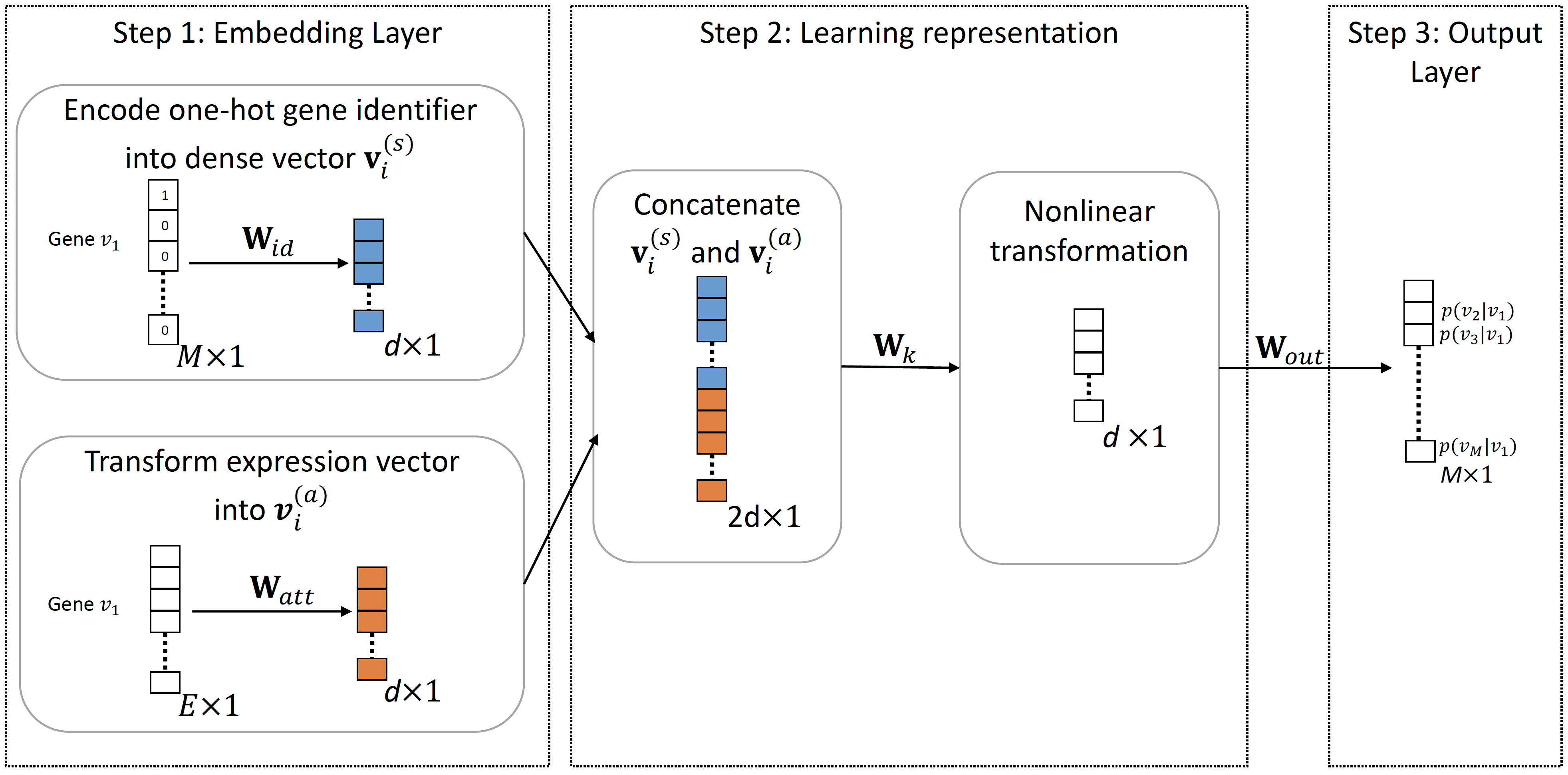
Architecture of GNE
Requirements
- TensorFlow (1.0 or later)
- python3.6
- sklearn
- networkx
- scipy
Example to run demo.
python run_GNE.py
Data
In order to use your own data, you have to provide
- an edgelist representing the interaction network which is converted to M by M adjacency matrix, and
- an M by E feature matrix (E is the number of experiments to measure gene expression)
We load interaction network data and expression data matrix for yeast in run_GNE.py as an example. The original datasets can be found from data sources:
| Dataset | Source |
|---|---|
| Interaction dataset | BioGRID |
| Gene expression data | DREAM5 Challenge |
You can use M by M adjacency matrix as input by editing run_GNE.py.
Note: the order of genes in adjacency matrix and feature matrix should be same.
Settings
You can choose between the following settings:
- GNE: Setting λ = 0, GNE learns from only topological properties
- GNE+: Setting λ = 1, GNE learns from integration of topological properties and expression data
For coding, We name λ as alpha in parameters list.
Supplementary Materials
Supplementary Table S1
Supplementary Table S2
For more details, visit GNE references.
Contact
kk3671@rit.edu
If you find this code useful, please cite us as:
@article{kishan2019gne,
title={GNE: a deep learning framework for gene network inference by aggregating biological information},
author={Kishan, KC and Li, Rui and Cui, Feng and Yu, Qi and Haake, Anne R},
journal={BMC systems biology},
volume={13},
number={2},
pages={38},
year={2019},
publisher={Springer}
}